The study was published in Nature Genetics.
In this study, the researchers used a set of chromosome single segment substitution lines derived from a cross between the weak-dormancy japonica rice cultivar Nipponbare and the strong-dormancy aus rice cultivar Kasalath to identify a gene, named as Seed Dormancy 6 (SD6), that contributes to rice seed dormancy.
The researchers found that SD6 and its interaction partner ICE2 antagonistically control rice seed dormancy by regulating abscisic acid (ABA) homeostasis. Specifically, SD6 directly promotes the expression of the ABA catabolism gene ABA8OX3 and indirectly inhibits the expression of the ABA biosynthetic gene NCED2, while ICE2 acts in an opposite manner.
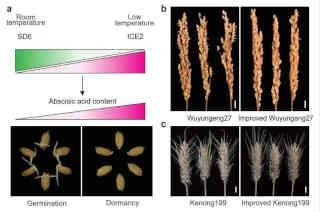
Temperature has a major effect on the strength of seed dormancy. The researchers revealed that the SD6/ICE2 molecular module controls rice seed dormancy in a temperature-dependent manner: SD6 is up-regulated to trigger seed germination when seeds are at room temperature. However, at low temperature, SD6 is down-regulated while ICE2 is up-regulated to maintain seed dormancy.
By editing SD6 in three rice cultivars, T619, Wu27, and Huai5, the researchers found that gene editing of SD6 could be a rapid and useful strategy for improving PHS tolerance in rice. Interestingly, editing the TaSD6 gene in the wheat variety Kenong199 also greatly improved PHS resistance in wheat, indicating that the SD6 gene is functionally conserved in controlling seed dormancy in both rice and wheat.
In summary, SD6 and ICE2 regulate seed dormancy by fine-tuning ABA content in seeds depending on temperature. In this way, they help seeds overcome natural seasonal change and ensure successful reproduction. For this reason, SD6 may be a powerful target for improving PHS resistance in cereals under field conditions.
Click here to see more...