Lateral branching in tomatoes is a complex genetic trait that directly impacts plant shape and overall productivity. Excessive branching can lead to competition for resources within the plant, ultimately reducing crop yields.
While numerous genes have been linked to branching traits, the intricate genetic network controlling this process has remained largely elusive. This research is a significant step toward unraveling this complexity, laying the groundwork for more optimized tomato plant architectures that could enhance agricultural efficiency.
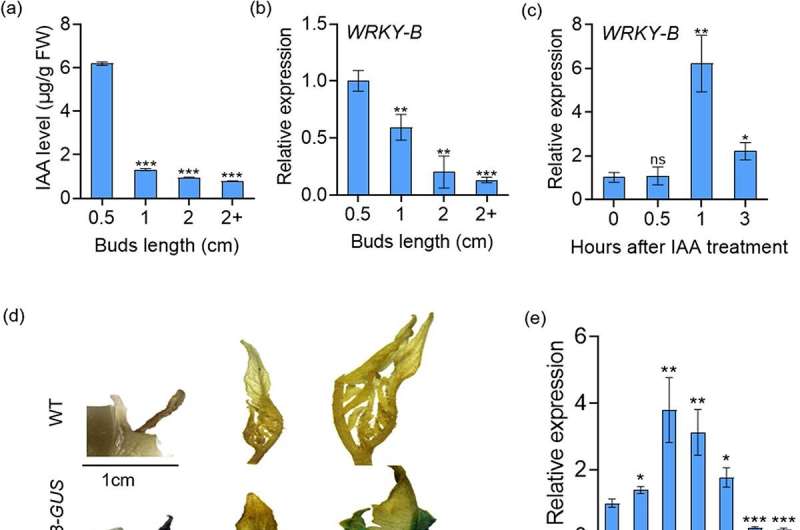
The study, published in Horticulture Research, was led by a team of researchers from China Agricultural University and Northeast Agricultural University. The scientists explored how the WRKY-B transcription factor interacts with key genes such as BLIND, PIN4, and IAA15, shedding light on how these interactions control lateral branch development in tomatoes.